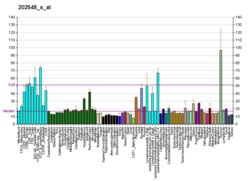
ARHGEF7
| ARHGEF7 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ARHGEF7, BETA-PIX, COOL-1, COOL1, Nbla10314, P50, P50BP, P85, P85COOL1, P85SPR, PAK3, PIXB, Rho guanine nucleotide exchange factor 7 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 605477; MGI: 1860493; HomoloGene: 2895; GeneCards: ARHGEF7; OMA:ARHGEF7 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Rho guanine nucleotide exchange factor 7 is a protein that in humans is encoded by the ARHGEF7 gene.[5][6][7][8]
ARHGEF7 is commonly known as the p21-activated protein kinase exchange factor beta (beta-PIX or βPIX), because it was identified by binding to p21-activated kinase (PAK) and also contains a guanine nucleotide exchange factor domain.[6]
Domains and functions
βPIX is a multidomain protein that functions both as a signaling scaffold protein and as an enzyme.[9] βPIX shares this domain structure and signaling function with the highly similar ARHGEF6/αPIX protein.
βPIX undergoes extensive alternative splicing to generate multiple variant proteins containing or lacking particular protein domains.[9] Adult forms all lack the amino terminal CH domain, and the two major adult variants have alternate carboxyl terminal region (termed β1 and β2): β1 forms contain the coiled-coil trimerization domain and the PDZ-target motif for binding to PDZ proteins (see below), while β2 forms lack both domains and their corresponding functions.[9]
βPIX contains a central DH/PH RhoGEF domain that functions as a guanine nucleotide exchange factor (GEF) for small GTPases of the Rho family, and specifically Rac and Cdc42.[6] Like other GEFs, βPIX can promote both release of GDP from an inactive small GTP-binding protein and binding of GTP to promote its activation. Signaling scaffolds bind to specific partners to promote efficient signal transduction by arranging sequential elements of a pathway near each other to facilitate interaction/information transfer, and also by holding these partner protein complexes in specific locations within the cell to promote local or regional signaling. In the case of βPIX, its SH3 domain binds to partner proteins with appropriate polyproline motifs, and particularly to group I p21-activated kinases (PAKs) (PAK1, PAK2 and PAK3).[6] PAK is bound to the βPIX SH3 domain in the inactive state, and activated Rac1 or Cdc42 binding to this PAK stimulates its protein kinase activity leading to downstream target protein phosphorylation; since βPIX can activate the "p21" small GTPases Rac1 or Cdc42 through its GEF activity, this βPIX/PAK/Rac complex exemplifies a scaffolding function.
Structurally, βPIX assembles as a trimer through a carboxyl-terminal coiled-coil domain that is present in the major carboxyl terminal splice variant β1, and further interacts with dimers of GIT1 or GIT2 through a nearby GIT-binding domain to form oligomeric GIT-PIX complexes.[9] Through this GIT-PIX complex, the scaffolding function of βPIX is amplified by also being able to hold GIT partners in proximity to βPIX partners. In contrast, β2 carboxyl terminal variants lack this coiled-coil region and are predicted to be unable to trimerize. The major carboxyl terminal variant β1 also has a PDZ domain binding target motif that binds to the PDZ domains in SHANK1,[10] scribble,[11] and SNX27[12] proteins. Some splice variants of βPIX contain an amino-terminal Calponin Homology (CH) domain whose functions remain relatively poorly defined, but may interacts with parvin/affixin family proteins. [13][9] βPIX variants with this extended amino terminal CH domain are most highly expressed early in development, but appear rare after birth.[9]
Interactions
βPIX has been reported to interact with over 120 proteins.[9][14]
Major interacting proteins include:
- Itself, or the highly-related ARHGEF6/αPIX via a trimeric coiled-coil interaction.
- GIT1 or GIT2 dimers via GIT-binding domain.
- p21-activated kinases (PAKs) 1, 2 and 3 via SH3 domain.
- c-Cbl via SH3 domain.
- Rho family GTP-binding protein family members Rac1 and Cdc42, activated via DHPH RhoGEF domain.
- The neuronal synapse adaptors SHANK1, SHANK2, and SHANK3 via PDZ
- Scribble via PDZ
- SNX27 via PDZ
See also
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000102606 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000031511 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Oh WK, Yoo JC, Jo D, Song YH, Kim MG, Park D (July 1997). "Cloning of a SH3 domain-containing proline-rich protein, p85SPR, and its localization in focal adhesion". Biochemical and Biophysical Research Communications. 235 (3): 794–798. doi:10.1006/bbrc.1997.6875. PMID 9207241.
- ^ a b c d Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L (July 1998). "PAK kinases are directly coupled to the PIX family of nucleotide exchange factors". Molecular Cell. 1 (2): 183–192. doi:10.1016/S1097-2765(00)80019-2. PMID 9659915.
- ^ Bagrodia S, Taylor SJ, Jordon KA, Van Aelst L, Cerione RA (October 1998). "A novel regulator of p21-activated kinases". Journal of Biological Chemistry. 273 (37): 23633–23636. doi:10.1074/jbc.273.37.23633. PMID 9726964.
- ^ "Entrez Gene: ARHGEF7 Rho guanine nucleotide exchange factor (GEF) 7".
- ^ a b c d e f g Zhou W, Li X, Premont RT (May 2016). "Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes". Journal of Cell Science. 129 (10): 1963–1974. doi:10.1242/jcs.179465. PMC 6518221. PMID 27182061.
- ^ Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E (2003). "The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42". J. Biol. Chem. 278 (21): 19220–9. doi:10.1074/jbc.M301052200. PMID 12626503.
- ^ Audebert S, Navarro C, Nourry C, Chasserot-Golaz S, Lecine P, Bellaiche Y, Dupont JL, Premont RT, Sempere C, Strub JM, Van Dorsselaer A, Vitale N, Borg JP (June 2004). "Mammalian Scribble forms a tight complex with the betaPIX exchange factor". Current Biology. 14 (11): 987–995. doi:10.1016/j.cub.2004.05.051. PMID 15182672.
- ^ Valdes JL, Tang J, McDermott MI, Kuo JC, Zimmerman SP, Wincovitch SM, Waterman CM, Milgram SL, Playford MP (November 2011). "Sorting nexin 27 protein regulates trafficking of a p21-activated kinase (PAK) interacting exchange factor (β-Pix)-G protein-coupled receptor kinase interacting protein (GIT) complex via a PDZ domain interaction". Journal of Biological Chemistry. 286 (45): 39403–39416. doi:10.1074/jbc.M111.260802. PMC 3234764. PMID 21926430.
- ^ Rosenberger G, Jantke I, Gal A, Kutsche K (2003). "Interaction of alphaPIX (ARHGEF6) with beta-parvin (PARVB) suggests an involvement of alphaPIX in integrin-mediated signaling". Human Molecular Genetics. 12 (2): 155–167. doi:10.1093/hmg/ddg019. PMID 12499396.
- ^ "ARHGEF7 Result Summary".
Further reading
- Turner CE, Brown MC, Perrotta JA, Riedy MC, Nikolopoulos SN, McDonald AR, Bagrodia S, Thomas S, Leventhal PS (1999). "Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: A role in cytoskeletal remodeling". J. Cell Biol. 145 (4): 851–63. doi:10.1083/jcb.145.4.851. PMC 2133183. PMID 10330411.
- Bagrodia S, Bailey D, Lenard Z, Hart M, Guan JL, Premont RT, Taylor SJ, Cerione RA (1999). "A tyrosine-phosphorylated protein that binds to an important regulatory region on the cool family of p21-activated kinase-binding proteins". J. Biol. Chem. 274 (32): 22393–400. doi:10.1074/jbc.274.32.22393. PMID 10428811.
- Premont RT, Claing A, Vitale N, Perry SJ, Lefkowitz RJ (2000). "The GIT family of ADP-ribosylation factor GTPase-activating proteins. Functional diversity of GIT2 through alternative splicing". J. Biol. Chem. 275 (29): 22373–80. doi:10.1074/jbc.275.29.22373. PMID 10896954.
- Ku GM, Yablonski D, Manser E, Lim L, Weiss A (2001). "A PAK1-PIX-PKL complex is activated by the T-cell receptor independent of Nck, Slp-76 and LAT". EMBO J. 20 (3): 457–65. doi:10.1093/emboj/20.3.457. PMC 133476. PMID 11157752.
- Koh CG, Tan EJ, Manser E, Lim L (2002). "The p21-activated kinase PAK is negatively regulated by POPX1 and POPX2, a pair of serine/threonine phosphatases of the PP2C family". Curr. Biol. 12 (4): 317–21. doi:10.1016/S0960-9822(02)00652-8. PMID 11864573.
- Brown MC, West KA, Turner CE (2002). "Paxillin-dependent paxillin kinase linker and p21-activated kinase localization to focal adhesions involves a multistep activation pathway". Mol. Biol. Cell. 13 (5): 1550–65. doi:10.1091/mbc.02-02-0015. PMC 111126. PMID 12006652.
- Shin EY, Shin KS, Lee CS, Woo KN, Quan SH, Soung NK, Kim YG, Cha CI, Kim SR, Park D, Bokoch GM, Kim EG (2003). "Phosphorylation of p85 beta PIX, a Rac/Cdc42-specific guanine nucleotide exchange factor, via the Ras/ERK/PAK2 pathway is required for basic fibroblast growth factor-induced neurite outgrowth". J. Biol. Chem. 277 (46): 44417–30. doi:10.1074/jbc.M203754200. PMID 12226077.
- Rosenberger G, Jantke I, Gal A, Kutsche K (2003). "Interaction of alphaPIX (ARHGEF6) with beta-parvin (PARVB) suggests an involvement of alphaPIX in integrin-mediated signaling". Hum. Mol. Genet. 12 (2): 155–67. doi:10.1093/hmg/ddg019. PMID 12499396.
- Mignone F, Grillo G, Liuni S, Pesole G (2003). "Computational identification of protein coding potential of conserved sequence tags through cross-species evolutionary analysis". Nucleic Acids Res. 31 (15): 4639–45. doi:10.1093/nar/gkg483. PMC 169873. PMID 12888525.
- Yamamoto Y, Fujimoto Y, Arai R, Fujie M, Usami S, Yamada T (2003). "Retrotransposon-mediated restoration of Chlorella telomeres: accumulation of Zepp retrotransposons at termini of newly formed minichromosomes". Nucleic Acids Res. 31 (15): 4646–53. doi:10.1093/nar/gkg490. PMC 169880. PMID 12888526.
- Flanders JA, Feng Q, Bagrodia S, Laux MT, Singavarapu A, Cerione RA (2003). "The Cbl proteins are binding partners for the Cool/Pix family of p21-activated kinase-binding proteins". FEBS Lett. 550 (1–3): 119–23. doi:10.1016/S0014-5793(03)00853-6. PMID 12935897. S2CID 46540220.
- Shin EY, Woo KN, Lee CS, Koo SH, Kim YG, Kim WJ, Bae CD, Chang SI, Kim EG (2004). "Basic fibroblast growth factor stimulates activation of Rac1 through a p85 betaPIX phosphorylation-dependent pathway". J. Biol. Chem. 279 (3): 1994–2004. doi:10.1074/jbc.M307330200. PMID 14557270.
- Lim CS, Kim SH, Jung JG, Kim JK, Song WK (2004). "Regulation of SPIN90 phosphorylation and interaction with Nck by ERK and cell adhesion". J. Biol. Chem. 278 (52): 52116–23. doi:10.1074/jbc.M310974200. PMID 14559906.
External links
- ARHGEF7 Info with links in the Cell Migration Gateway
- Human ARHGEF7 genome location and ARHGEF7 gene details page in the UCSC Genome Browser.
- Human PAK3 genome location and PAK3 gene details page in the UCSC Genome Browser.
- v
- t
- e
-
 1by1: DBL homology domain from beta-PIX
1by1: DBL homology domain from beta-PIX -
 1zsg: beta PIX-SH3 complexed with an atypical peptide from alpha-PAK
1zsg: beta PIX-SH3 complexed with an atypical peptide from alpha-PAK -
 2ak5: beta PIX-SH3 complexed with a Cbl-b peptide
2ak5: beta PIX-SH3 complexed with a Cbl-b peptide -
 2df6: Crystal Structure of the SH3 Domain of betaPIX in Complex with a High Affinity Peptide from PAK2
2df6: Crystal Structure of the SH3 Domain of betaPIX in Complex with a High Affinity Peptide from PAK2 -
 2esw: Atomic structure of the N-terminal SH3 domain of mouse beta PIX,p21-activated kinase (PAK)-interacting exchange factor
2esw: Atomic structure of the N-terminal SH3 domain of mouse beta PIX,p21-activated kinase (PAK)-interacting exchange factor -
 2g6f: Crystal Structure of the SH3 Domain of betaPIX in Complex with a High Affinity Peptide from PAK2
2g6f: Crystal Structure of the SH3 Domain of betaPIX in Complex with a High Affinity Peptide from PAK2
 | This article on a gene on human chromosome 13 is a stub. You can help Wikipedia by expanding it. |
- v
- t
- e